Tools & Models
RODES, a RObust DEsign Synthesis tool
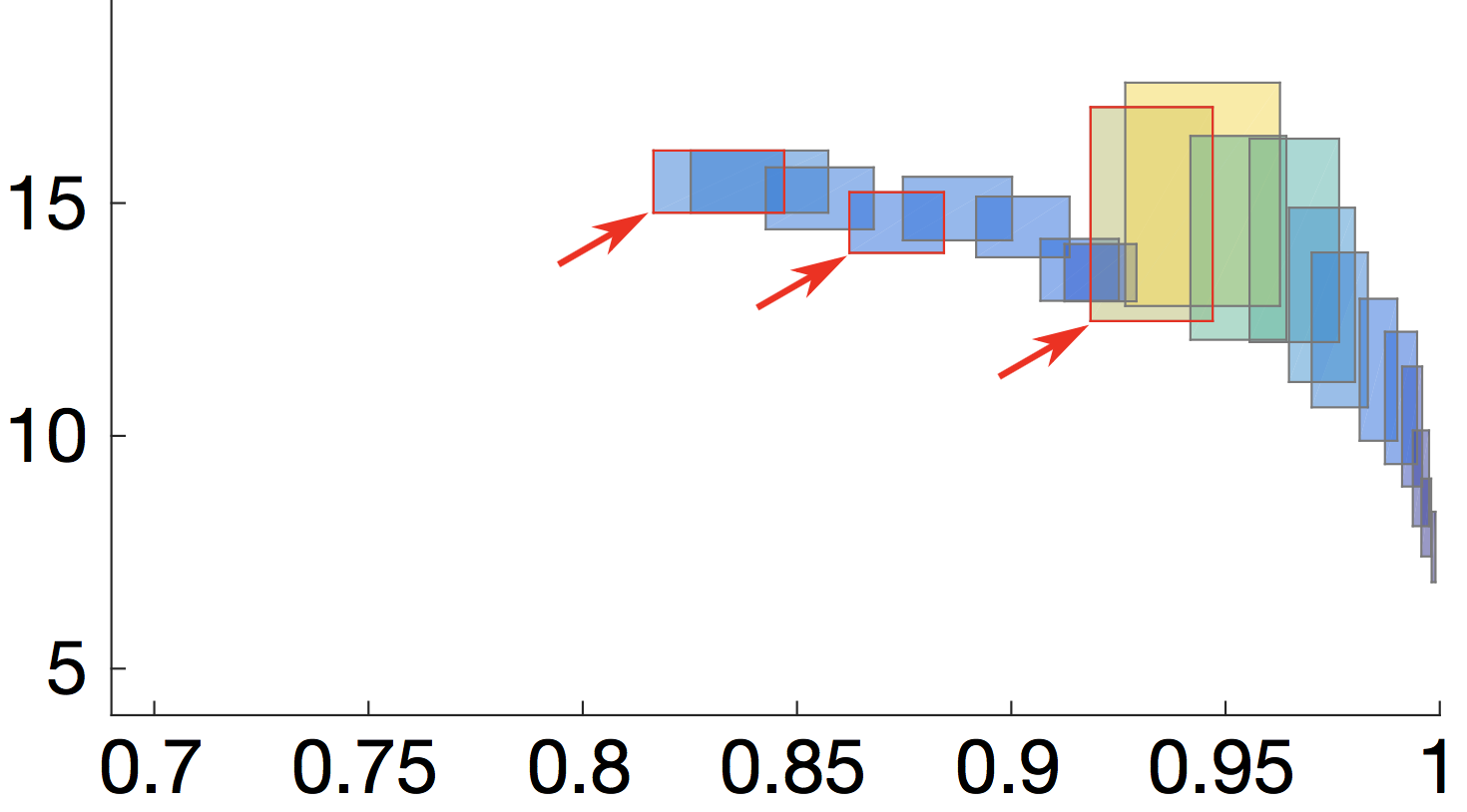
RODES is a tool for the synthesis of optimal and robust designs for probabilistic systems. Synthesized designs (given as parametric Markov chains) are 1) robust to pre-specified levels of variation in the system parameters; 2) satisfy strict performance, reliability and other quality constraints; and 3) are Pareto optimal with respect to a set of quality optimisation criteria.
The resulting Pareto front consists of quality attribute regions induced by the parametric designs (see picture). The size and shape of these regions provide key insights into the system robustness since they capture the sensitivity of quality attributes to parameter changes (i.e, small regions = robust designs).
The method is based on a combination of GPU-accelerated parameter synthesis for Markov chains (to compute the quality attribute regions for fixed discrete parameters) through the PRISM-PSY tool, and multi-objective optimization based on an extended, sensitivity-aware dominance relation. RODES further extends the PRISM language with constructs to enable the specification of parametric models.
HeartVerify: Model-Based Quantitative Verification of Cardiac Pacemakers
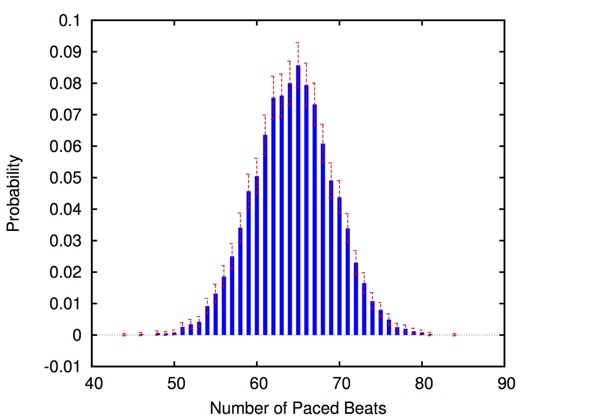
HeartVerify is a framework for the analysis and verification of pacemaker software and personalised heart models. Models are specified in MATLAB Stateflow and are analysed using the Cosmos tool for statistical model checking, thus enabling the analysis of complex nonlinear dynamics of the heart, where precise (numerical) verification methods typically fail.
HeartVerify is modular in that it allows configuring and testing different heart and pacemaker models in a plug-and-play fashion, without changing their communication interface or the verification engine. It supports the verification of probabilistic properties, together with additional analyses such as simulation, generation of probability density plots (see figure) and parametric analyses. HeartVerify comes with different heart and pacemaker models, including methods for model personalization from ECG data and rate-adaptive pacing.
PRISM-PSY: precise GPU-accelerated parameter synthesis for stochastic systems
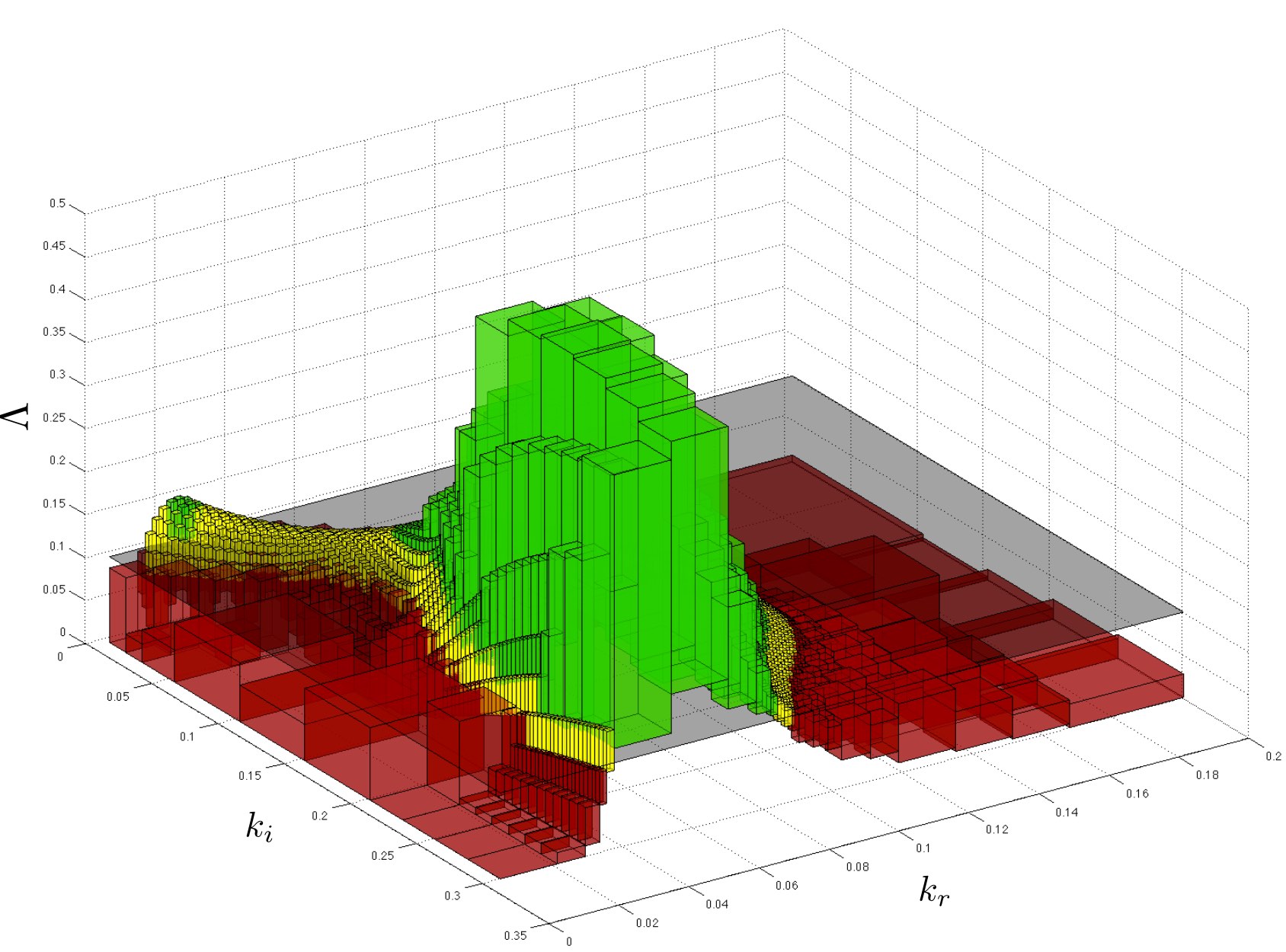
PRISM-PSY is a tool for precise GPU-accelerated parameter synthesis for continuous-time Markov chains and time-bounded temporal logic specifications (CSL). It finds parameter values such that a CSL property is guaranteed to hold, or, in the case of quantitative properties, the probability of satisfying the property is maximised or minimised.
The method builds on a parameteric extension of probabilistic model checking to compute probability bounds (formulated as matrix-vector operations for effective GPU acceleration) and refinement and sampling of the parameter space.
PRISM-PSY has been applied to a variety of biological and engineered systems, including models of molecular walkers, mammalian cell cycle, and the Google file system.
An integrated model of ecological and metabolic networks for bioremediation of contaminated food webs
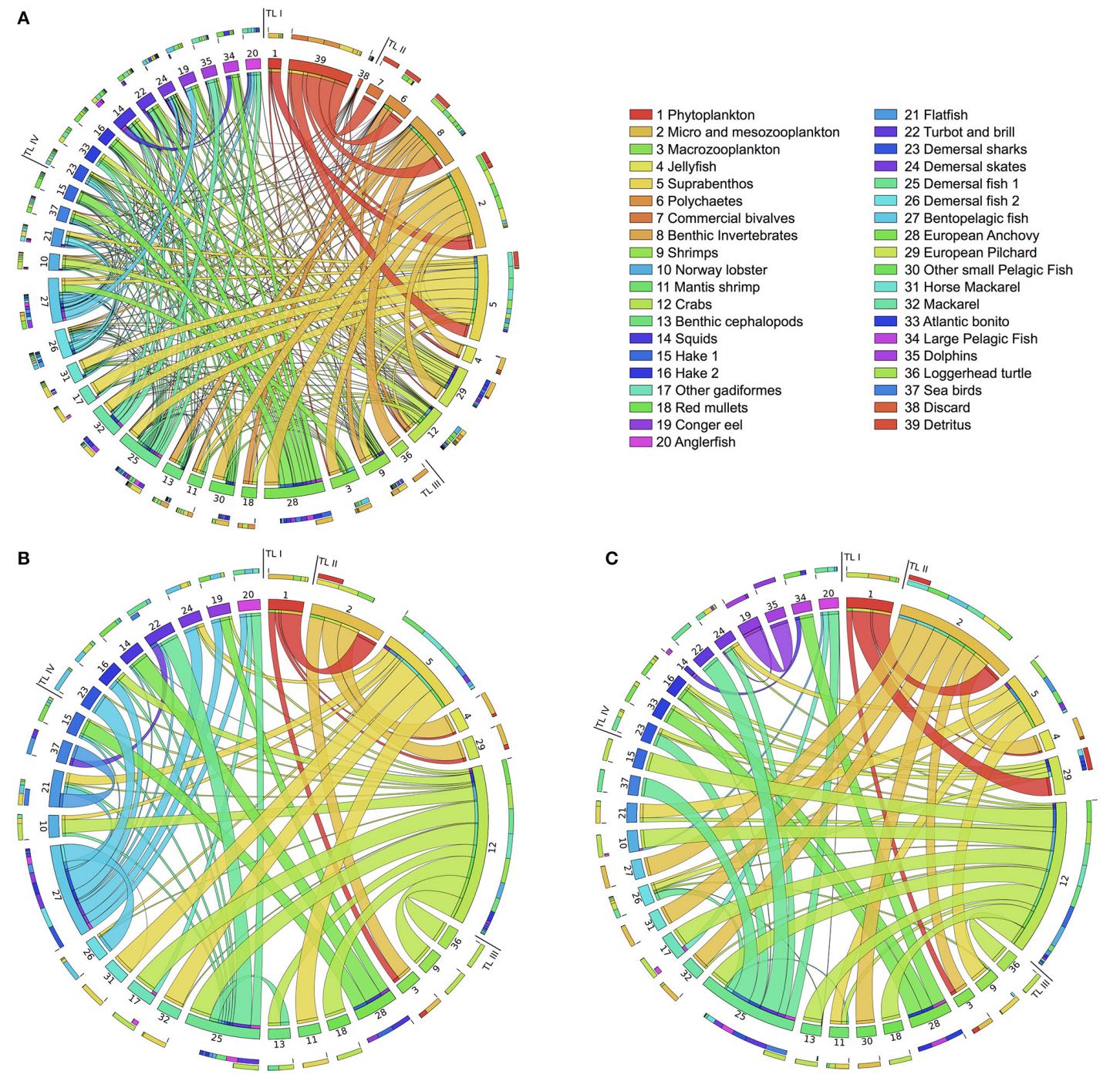
In this work we develop computational methods and models to study pollution dynamics in ecological networks and to shed light on three key questions: How persistent organic pollutants (e.g. PCBs that bind to the fat tissue) are transferred in food webs through feeding connections? What are the species that play role in the pollutant distribution? How to synthesize effective bioremediation strategies mediated by pollutant-degrading bacteria?
We present a computational framework to 1) reconstruct bioaccumulation networks from (partial) data; 2) identify key species in contamination dynamics through a new network index based on sensitivity analysis; and 3) analyze the multiscale effects of microbial bioremediation on species-level contamination by integrating metabolic networks of biodegrading bacteria and bioaccumulation networks.
We consider the case of PCBs bioaccumulation in the Adriatic food web and aerobic PCBs bioremediation by a strain of P. putida (data and models are included in the package).
IDHP heart model and personalization from ECG
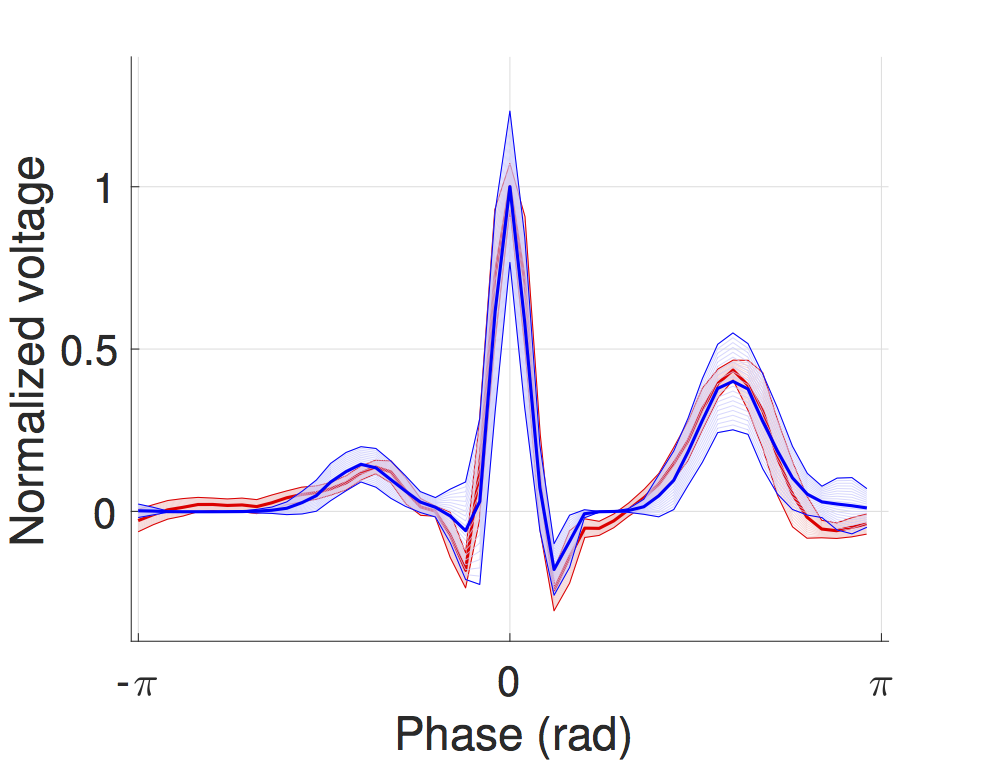
This model is a timed automata translation of the IDHP (Integrated Dual-chamber Heart and Pacer) model by Lian et al. Timed components realize the conduction delays between functional components of the heart and action potential propagation between nodes is implemented through synchronisation between the involved components. In this way, the model can be easily extended with other accessory conduction pathways in order to reproduce specific heart conditions.
The IDHP model can be also parametrised from patient electrocardiogram (ECG) in order to reproduce the specific physiological characteristics of the individual. For this purpose, we extended the model in order to generate synthetic ECG signals that reflect the heart events occurring at simulation time. Starting from raw ECG recordings, our method automatically finds model parameters such that the synthetic signal best mimics the input ECG, by minimising the statistical distance between the two. The resulting parameters correspond to probabilistic delays in order to reflect variability of ECG features.
The package below includes the IDHP heart model and its composition with the dual-chamber pacemaker model of Jiang et al. Personalization from ECG data is implemented in the HeartVerify tool.
Stochastic agent-based simulator for bone remodelling
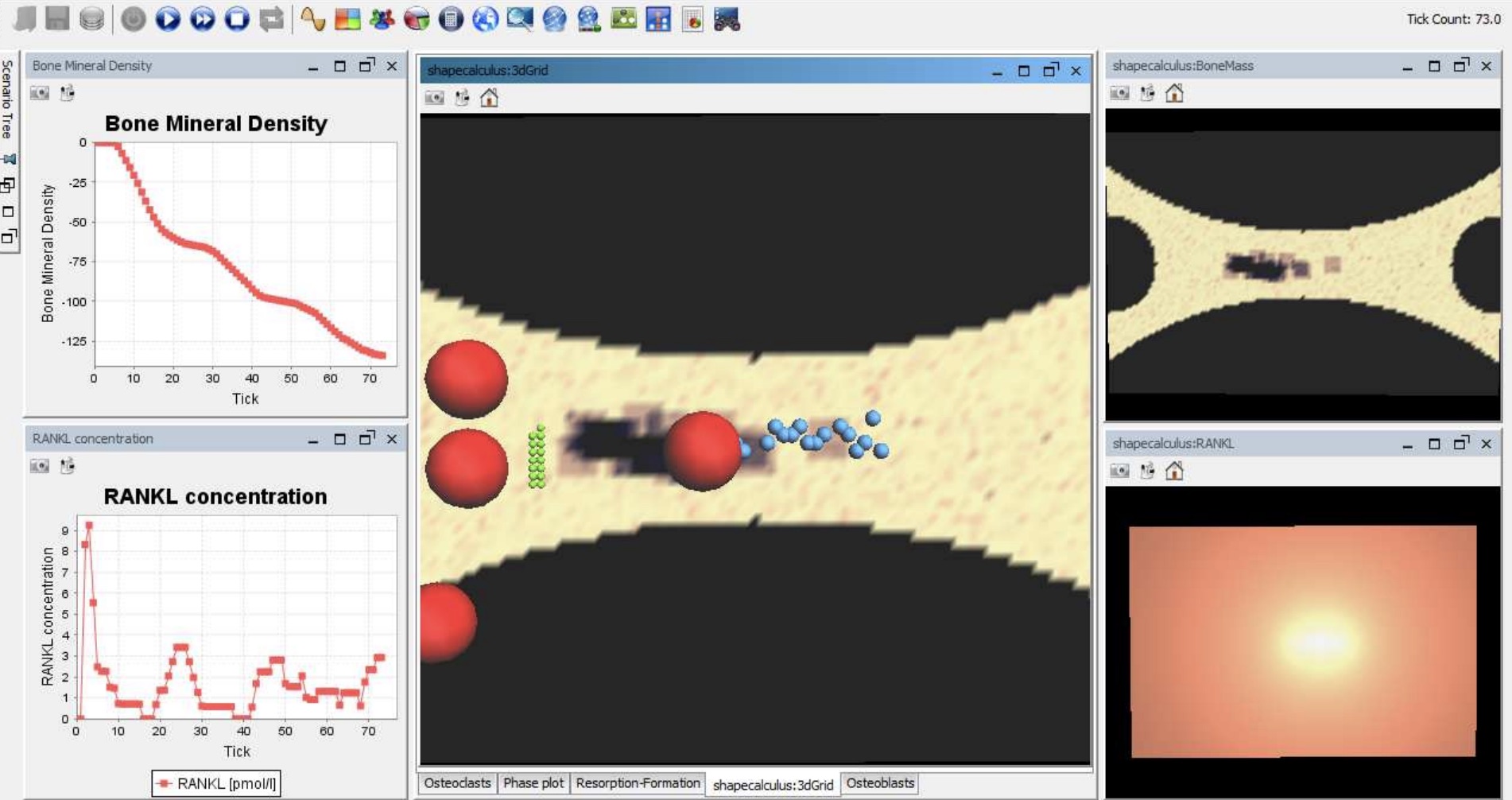
This is the first stochastic agent-based simulator for bone remodelling, i.e., the biological process of bone renewal. Bone remodelling is a paradigm for many homeostatic processes in the human body and consists of two highly coordinated phases: resorption of old bone by cells called osteoclasts, and formation of new bone by osteoblasts.
This tool builds on a multi-level modelling framework where agents are described at a higher level using the formal language of Shape Calculus, a spatial process algebra. Spatial aspects such as cell localisation and molecular gradients are indeed crucial in bone remodelling. We define a semantics in terms of stochastic agent-based models for the algebra and implement it in the Repast Simphony tool. The tool enables the simulation and visualization of bone cells, bone density and RANKL concetration affecting signalling, and can be parametrized in order to reproduce both healthy and osteoporotic conditions.
