Research
Probabilistic Robustness of Bayesian Neural Networks
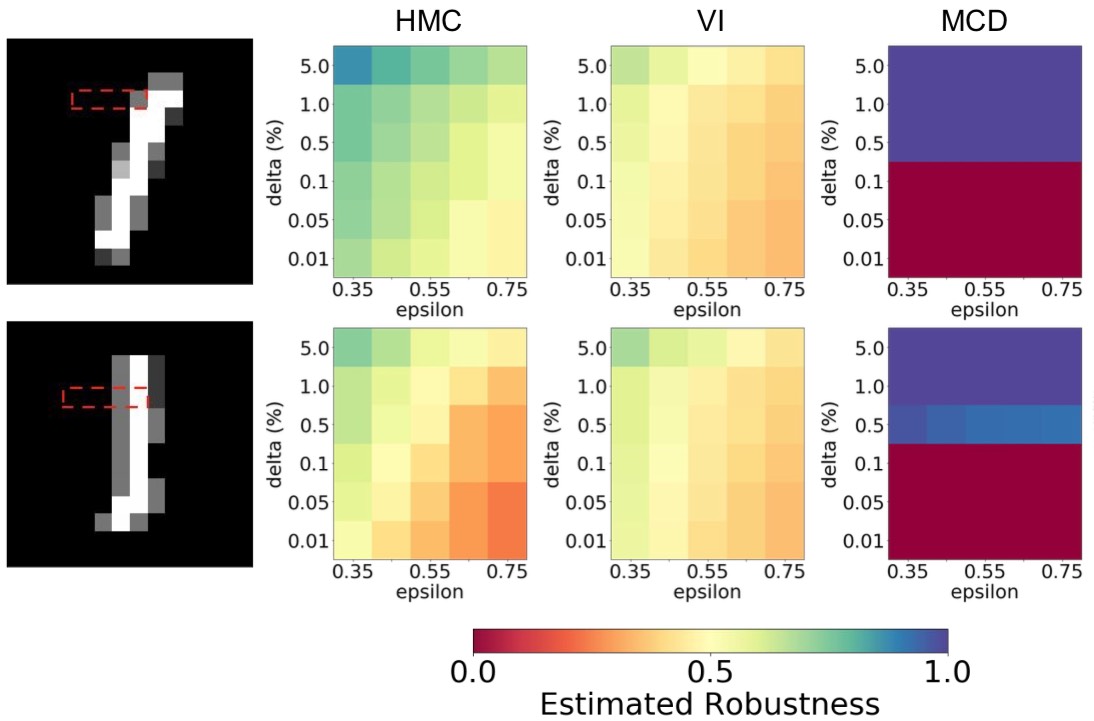
We introduce a probabilistic robustness measure for Bayesian Neural Networks (BNNs), defined as the probability that, given a test point, there exists a point within a bounded set such that the BNN prediction differs between the two. Such a measure can be used, for instance, to quantify the probability of the existence of adversarial examples. Building on statistical verification techniques for probabilistic models, we develop a framework that allows us to estimate probabilistic robustness for a BNN with statistical guarantees, i.e., with a priori error and confidence bounds, by leveraging a sequential probability estimation scheme based on the Massart bounds.
We evaluate our method with several approximate BNN inference techniques on image classification tasks associated to the MNIST digit recognition dataset and a two-class subset of the German Traffic Sign Recognition Benchmark (GTSRB) dataset, showing that the method enables quantification of uncertainty of BNN predictions in adversarial settings. Code available in GitHub.
Data-driven robust control for insulin therapy
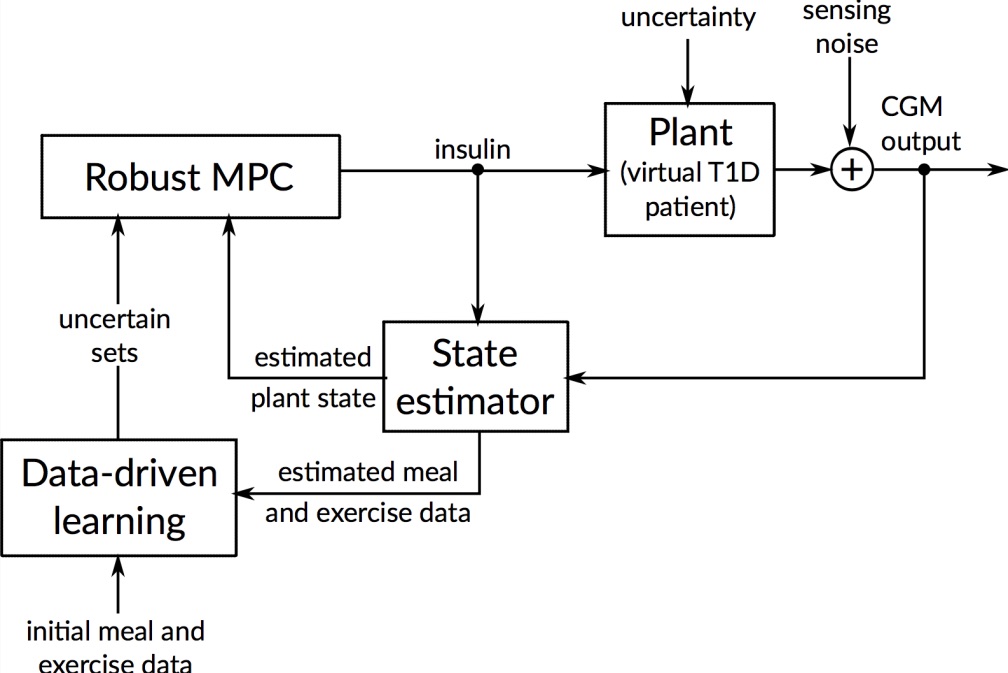
The artificial pancreas aims to automate treatment of type 1 diabetes (T1D) by integrating insulin pump and glucose sensor through control algorithms. However, fully closed-loop therapy is challenging since the blood glucose levels to control depend on disturbances related to the patient behavior, mainly meals and physical activity.
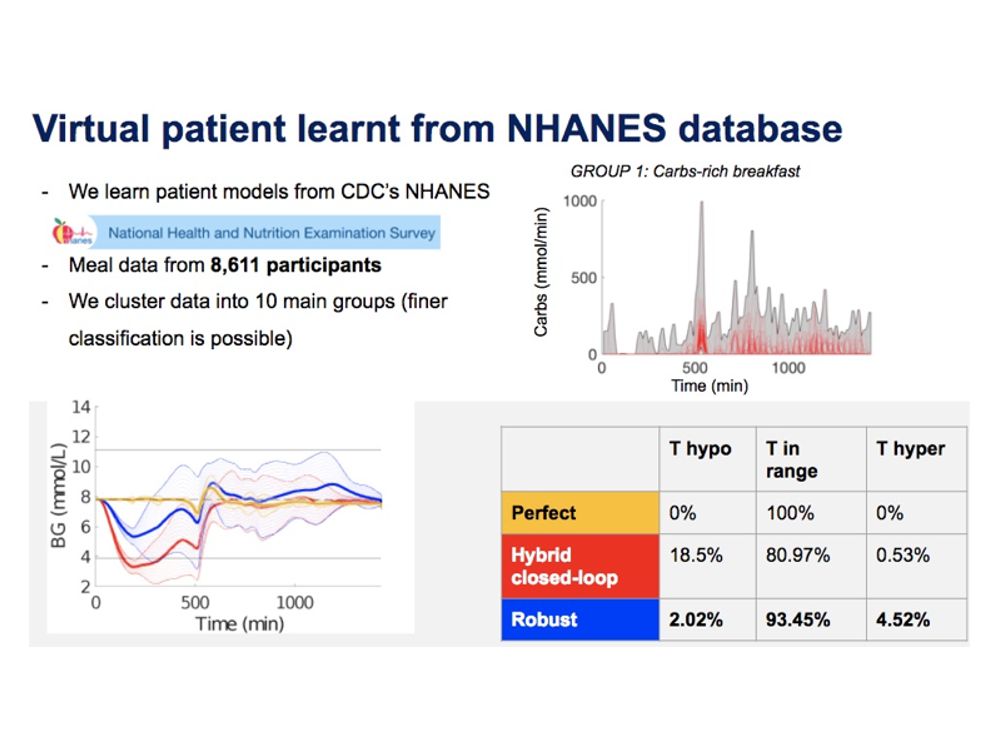
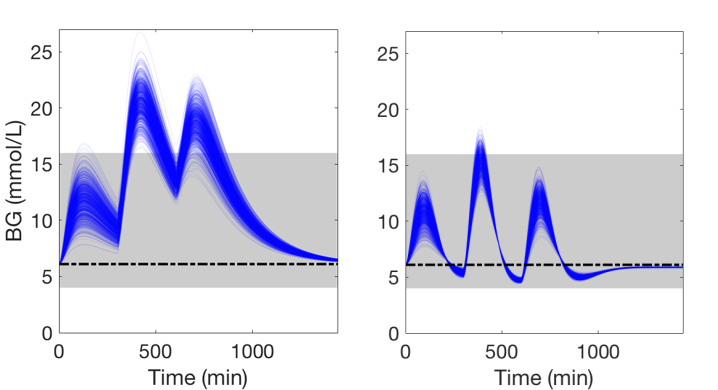
To handle meal and exercise uncertainties, in our work we construct data-driven models of meal and exercise behavior, and develop a robust model-predictive control (MPC) system able to reject such uncertainties, in this way eliminating the need for meal announcements by the patient. The data-driven models, called uncertainty sets, are built from data so that they cover the underlying (unknown) distribution with prescribed probabilistic guarantees. Then, our robust MPC system computes the insulin therapy that minimizes the worst-case performance with respect to these uncertainty sets, so providing a principled way to deal with uncertainty. State estimation follows a similar principle to MPC and exploits a prediction model to find the most likely state and disturbance estimate given the observations.
We evaluate our design on synthetic scenarios, including high-carbs intake and unexpected meal delays, and on large clusters of virtual patients learned from population-wide survey data sets (CDC NHANES).
SMT-based synthesis of safe and robust digital controllers
We present a new method for the automated synthesis of digital controllers with formal safety guarantees for systems with nonlinear dynamics, noisy output measurements, and stochastic disturbances. Our method derives digital controllers such that the corresponding closed-loop system, modeled as a sampled-data stochastic control system, satisfies a safety specification with probability above a given threshold. The method alternates between two steps: generation of a candidate controller (sub-optimal but rapid to generate), and verification of the candidate. The candidate is found by maximizing a Monte Carlo estimate of the safety probability, and by simulating the system with a non-validated ODE solver. To rule out unstable candidate controllers, we prove and utilize Lyapunov indirect method for instability of sampled-data nonlinear systems. In the verification step, we use a validated solver based on SMT to compute a numerically and statistically valid confidence interval for the safety probability of the candidate controller. If such probability is not above the threshold, we expand the search space for candidates by increasing the controller degree. We evaluate our technique on three case studies: an artificial pancreas model, a powertrain control model, and a quadruple-tank process.
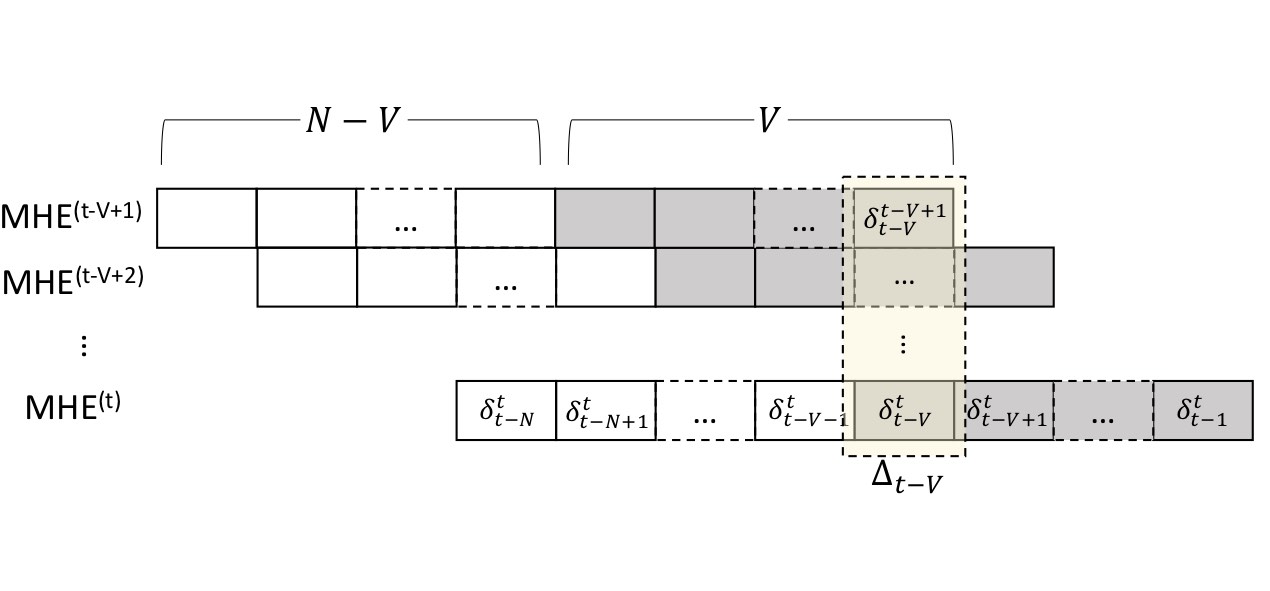
Committed Moving Horizon Estimation for Meal Detection
We introduce Committed Moving Horizon Estimation (CHME), a model-based technique for detecting and estimating unknown random disturbances in control systems. We investigate CHME in the context of meal detection and estimation method for the treatment of type 1 diabetes, where we are interested in automatically detecting the occurrence and estimate the amount of carbohydrate (CHO) intake from glucose sensor data. Meal detection and estimation is crucial in closed-loop insulin control as it can eliminate the need for manual meal announcements by the patient.
CMHE extends Moving Horizon Estimation, which alone, is not well-suited for disturbance estimation and meal detection. Indeed, accurate disturbance estimation needs both look-ahead (awareness of the disturbance effect on future system outputs) and history (past outputs to discriminate the start of the disturbance). For this purpose, CMHE aggregates the meal disturbances estimated by multiple MHE instances to balance future and past information at decision time, thus providing timely detection and accurate estimation. Applied to the detection of random meals from glucose sensor data, our method achieves an 88.5% overall detection rate and a 100% detection rate for the main meals (i.e., excluding snacks).
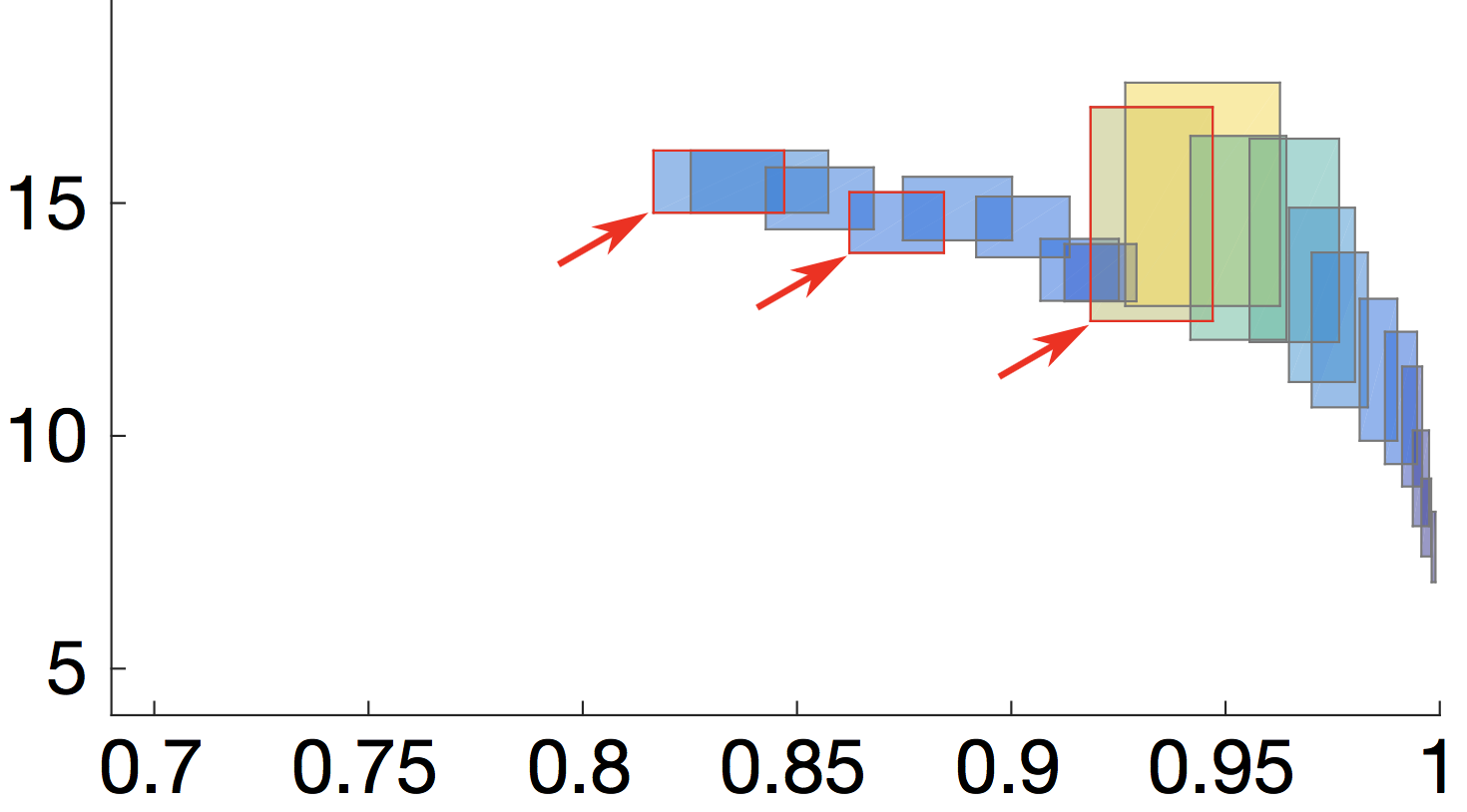
Robust design synthesis for probabilistic systems
We consider the problem of synthesizing optimal and robust designs (given as parametric Markov chains) that are 1) robust to pre-specified levels of variation in the system parameters; 2) satisfy strict performance, reliability and other quality constraints; and 3) are Pareto optimal with respect to a set of quality optimisation criteria.
The resulting Pareto front consists of quality attribute regions induced by the parametric designs (see picture). The size and shape of these regions provide key insights into the system robustness since they capture the sensitivity of quality attributes to parameter changes (i.e, small regions = robust designs).
The method is implemented in the RODES tool and is based on a combination of GPU-accelerated parameter synthesis for Markov chains (to compute the quality attribute regions for fixed discrete parameters) through the PRISM-PSY tool, and multi-objective optimization based on an extended, sensitivity-aware dominance relation.
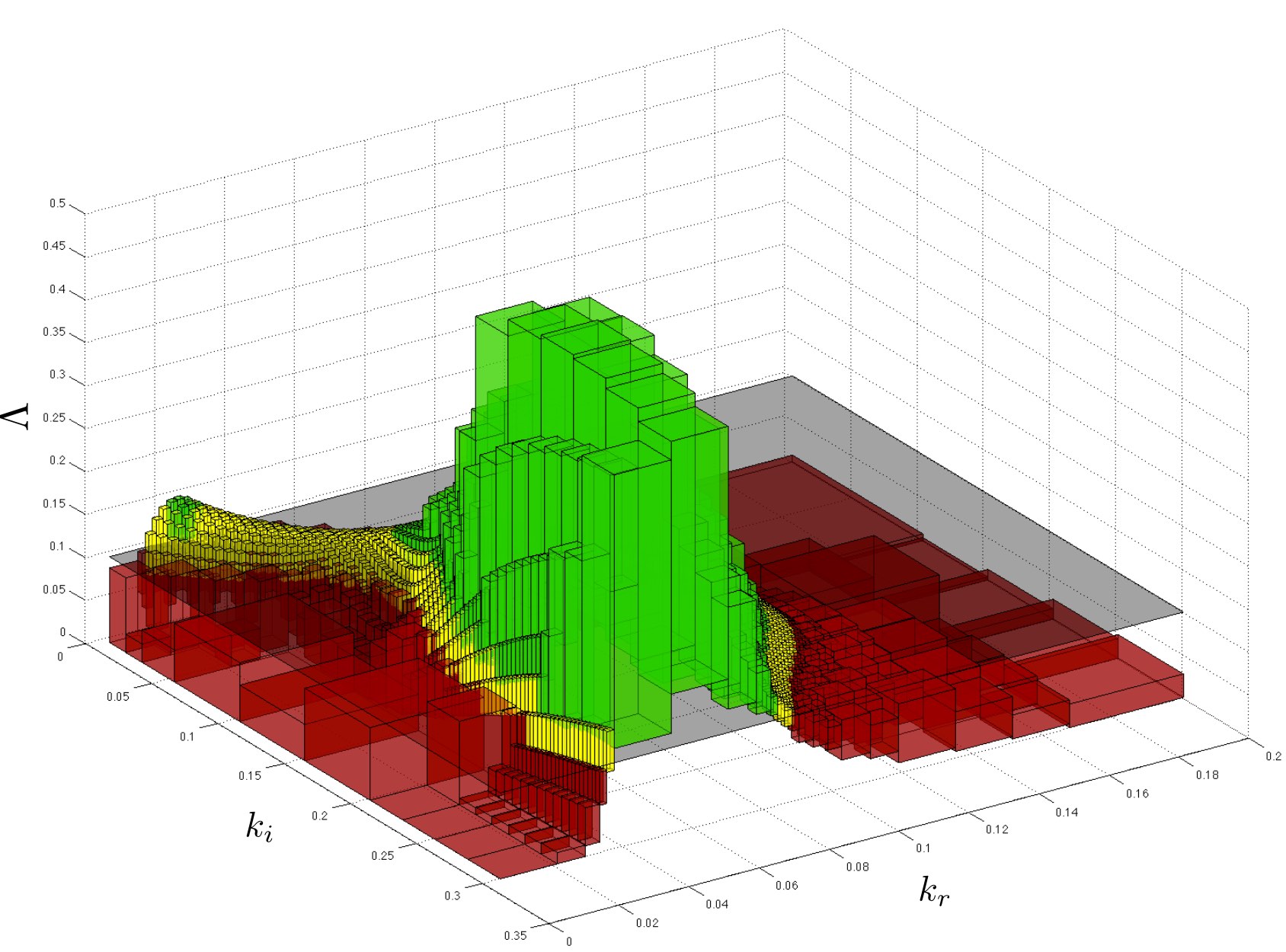
Precise parameter synthesis for continuous-time Markov chains
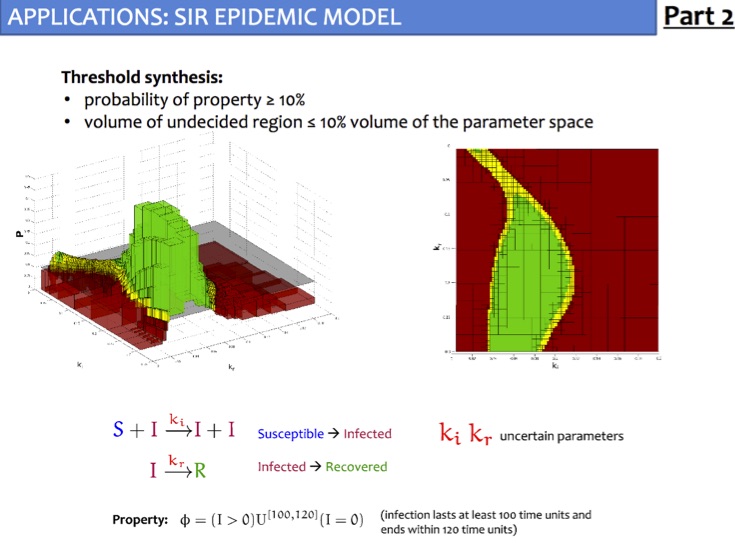
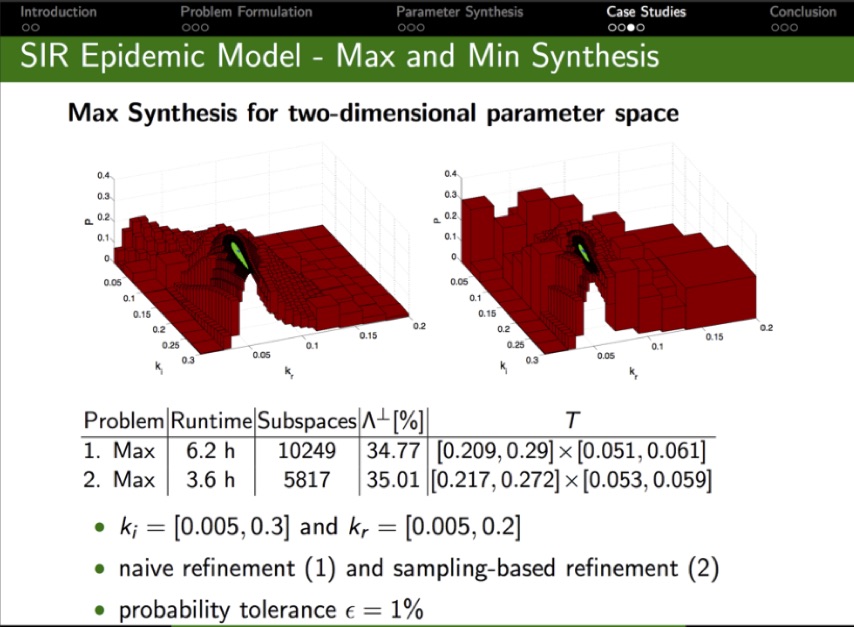
Given a parameteric continuous-time Markov chain (pCTMC), we aim at finding parameter values such that a CSL property is guaranteed to hold (threshold synthesis), or, in the case of quantitative properties, the probability of satisfying the property is maximised or minimised (max synthesis).
The solution of the threshold synthesis (see picture) is a decomposition of the parameter space into true and false regions that are guaranteed to, respectively, satisfy and violate the property, plus an arbitrarily small undecided region. On the other hand, in the max synthesis problem we identify an arbitrarily small region guaranteed to contain the actual maximum/minimum.
We developed synthesis methods based on a parameteric extension of probabilistic model checking to compute probability bounds, as well as refinement and sampling of the parameter space. We implemented GPU-accelerated algorithms for synthesis in the PRISM-PSY tool, which we applied to a variety of biological and engineered systems, including models of molecular walkers, mammalian cell cycle, and the Google file system.
Optimal synthesis of stochastic chemical reaction networks
The automatic derivation of Chemical Reaction Networks (CRNs) with prescribed behavior is one of the holy grails of synthetic biology, allowing for design automation of molecular devices and in the construction of predictive biochemical models.
In this work, we provide the first method for optimal syntax-guided synthesis of stochastic CRNs that is able to derive not just rate parameters but also the structure of the network. Borrowing from the programming language community, we propose a sketching language for CRNs that allows to specify the network as a partial program, using holes and variables to capture unknown components. Under the Linear Noise Approximation of the chemical master equation, a CRN sketch has a semantics in terms of parametric Ordinary Differential Equations (ODEs).
We support rich correctness properties that describe the temporal profile of the network as constraints over mean and variance of chemical species, and their higher order derivatives. In this way, we can synthesize networks where e.g., one of the species exhibit a bell-shape profile or has variance greater than its expectation.
We synthesize CRNs that satisfies the sketch constraints and a correctness specification while minimizing a given cost function (capturing the structural complexity of the network). The optimal synthesis algorithm employs SMT solvers over reals and ODEs (iSAT) and a meta-sketching abstraction that speeds up the search through cost constraints.
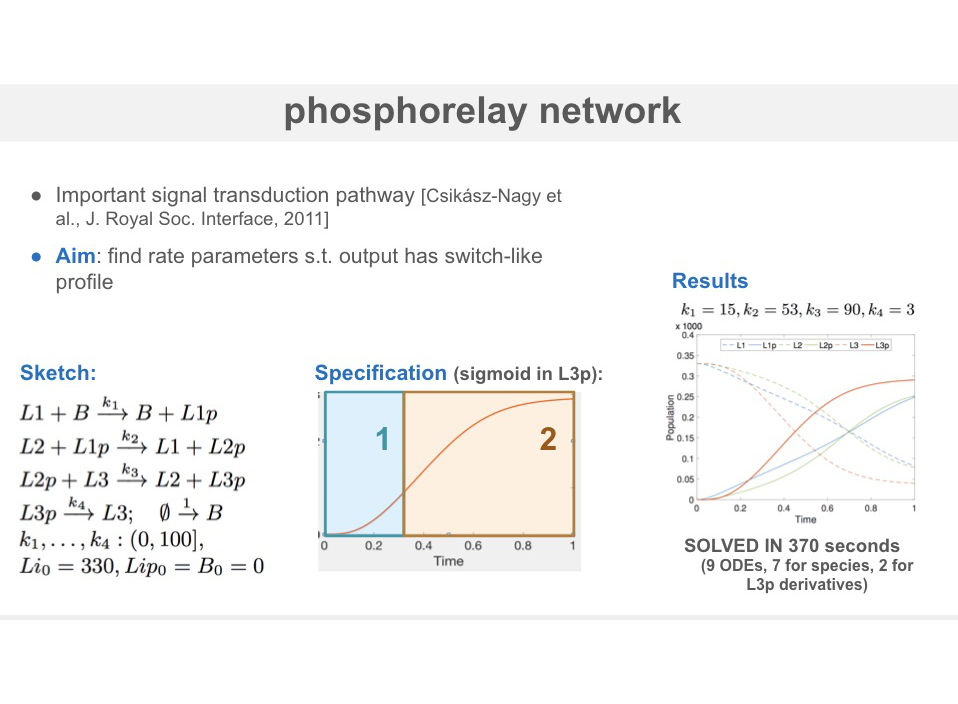
We evaluate the approach on a three key problems: synthesis of networks with a bell-shaped profile (occurring in signaling cascades), a CRN implementation of a stochastic process with prescribed levels of noise and synthesis of sigmoidal profiles in the phosphorelay network.
Closed-loop quantitative verification of rate-adaptive pacemakers
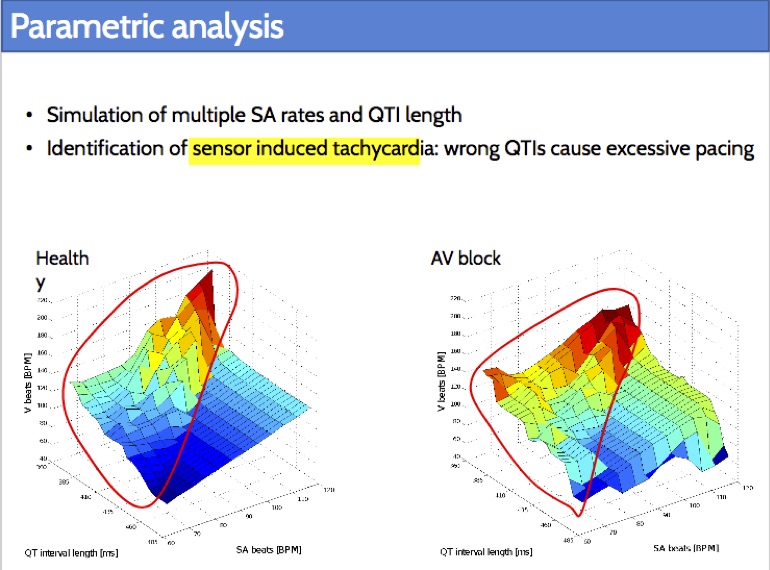
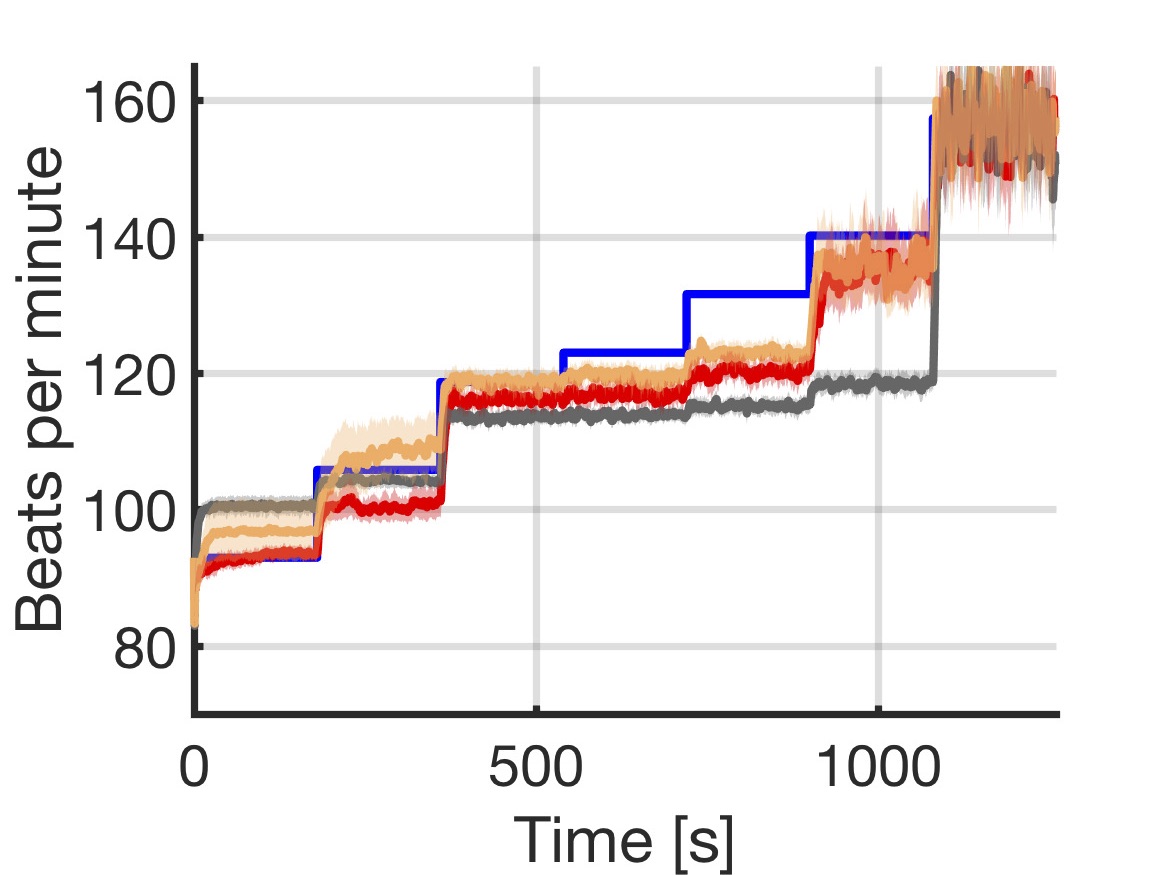
Rate-adaptive (RA) pacemakers are able to adjust the pacing rate depending on the levels of activity of the patient, detected by processing physiological signals data. RA pacemakers represent the only choice when the heart rate cannot naturally adapt to increasing demand (e.g. AV block). RA parameters depend on many patient-specific factors, and effective personalisation of such treatments can only be achieved through extensive exercise testing, which is normally intolerable for a cardiac patient.
We introduce a data-driven and model-based approach for the quantitative verification of RA pacemakers and formal analysis of personalised treatments. We developed a novel dual-sensor pacemaker model where the adaptive rate is computed by blending information from an accelerometer, and a metabolic sensor based on the QT interval (a feature of the ECG). The approach builds on the HeartVerify tool to provide statistical model checking of the probabilistic heart-pacemaker models, and supports model personalization from ECG data (see heart model page). Closed-loop analysis is achieved through the online generation of synthetic, model-based QT intervals and acceleration signals.
We further extend the model personalization method to estimate parameters from patient population, thus enabling safety verification of the device. We evaluate the approach on three subjects and a pool of virtual patients, providing rigorous, quantitative insights into the closed-loop behaviour of the device under different exercise levels and heart conditions.
HeartVerify: Model-Based Quantitative Verification of Cardiac Pacemakers
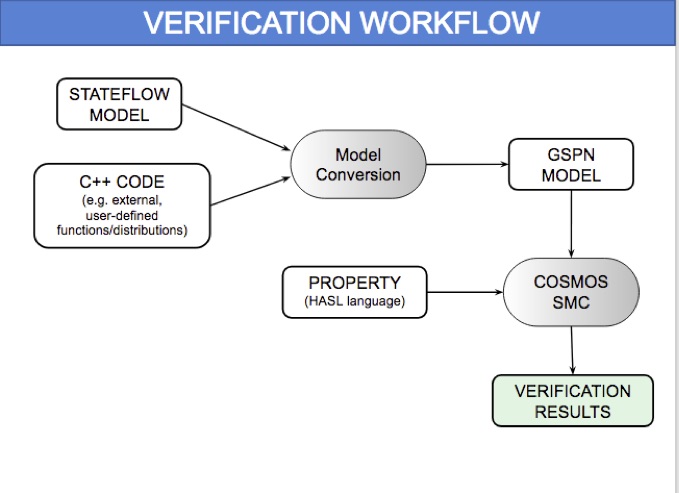
HeartVerify is a framework for the analysis and verification of pacemaker software and personalised heart models. Models are specified in MATLAB Stateflow and are analysed using the Cosmos tool for statistical model checking, thus enabling the analysis of complex nonlinear dynamics of the heart, where precise (numerical) verification methods typically fail.
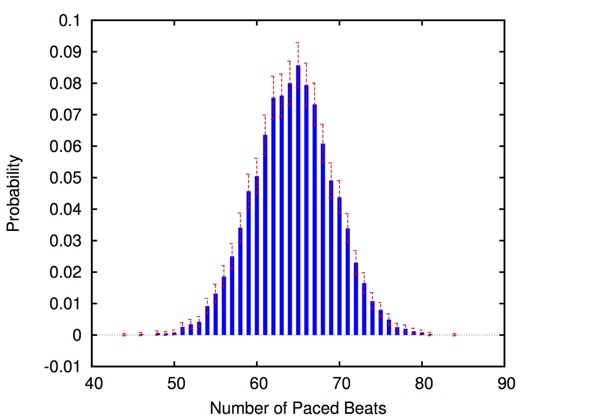
The approach is modular in that it allows configuring and testing different heart and pacemaker models in a plug-and-play fashion, without changing their communication interface or the verification engine. It supports the verification of probabilistic properties, together with additional analyses such as simulation, generation of probability density plots (see figure) and parametric analyses. HeartVerify comes with different heart and pacemaker models, including methods for model personalization from ECG data and rate-adaptive pacing.
Probabilistic timed modelling of cardiac dynamics and personalization from ECG
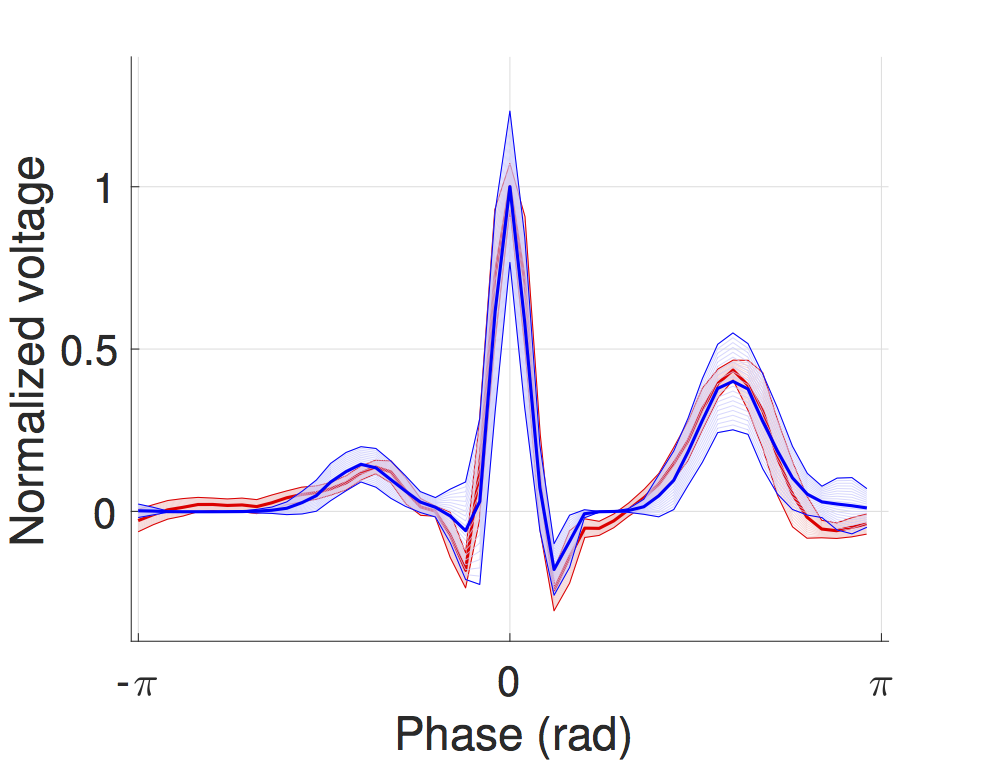
We developed a timed automata translation of the IDHP (Integrated Dual-chamber Heart and Pacer) model by Lian et al. Timed components realize the conduction delays between functional components of the heart and action potential propagation between nodes is implemented through synchronisation between the involved components. In this way, the model can be easily extended with other accessory conduction pathways in order to reproduce specific heart conditions.
The IDHP model can be also parametrised from patient electrocardiogram (ECG) in order to reproduce the specific physiological characteristics of the individual. For this purpose, we extended the model in order to generate synthetic ECG signals that reflect the heart events occurring at simulation time. Starting from raw ECG recordings, our method automatically finds model parameters such that the synthetic signal best mimics the input ECG, by minimising the statistical distance between the two. The resulting parameters correspond to probabilistic delays in order to reflect variability of ECG features.
The IDHP heart model can be downloaded from the tool page, while personalization from ECG data is implemented in the HeartVerify tool.
Formal analysis of bone remodelling
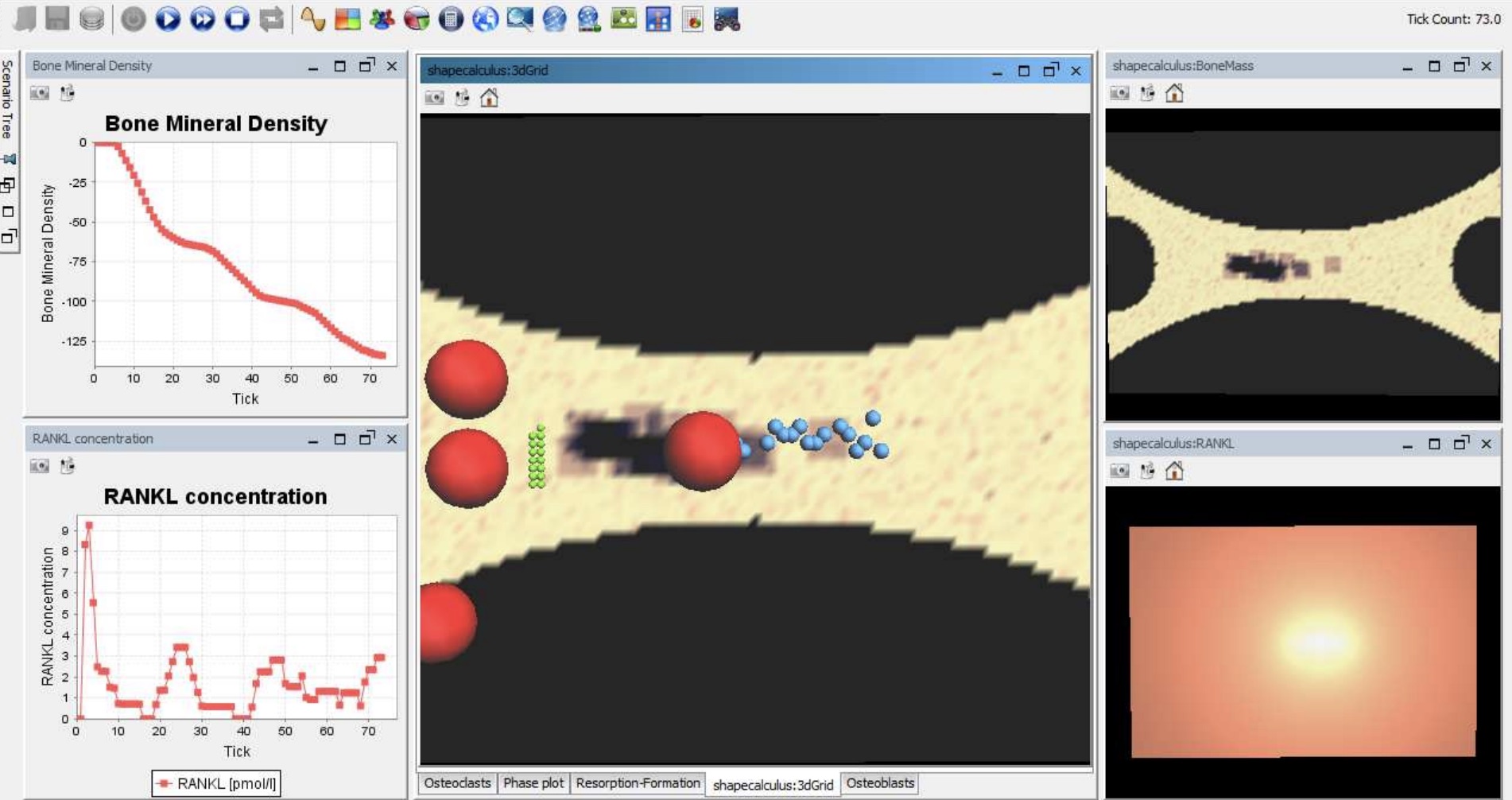
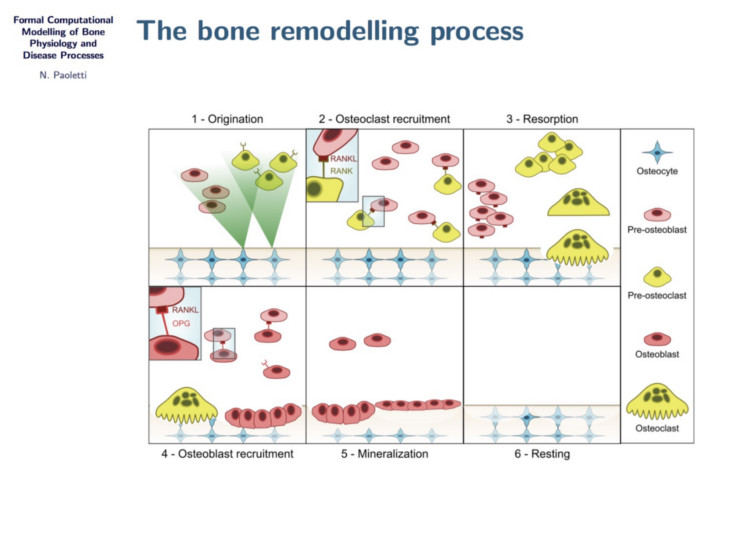
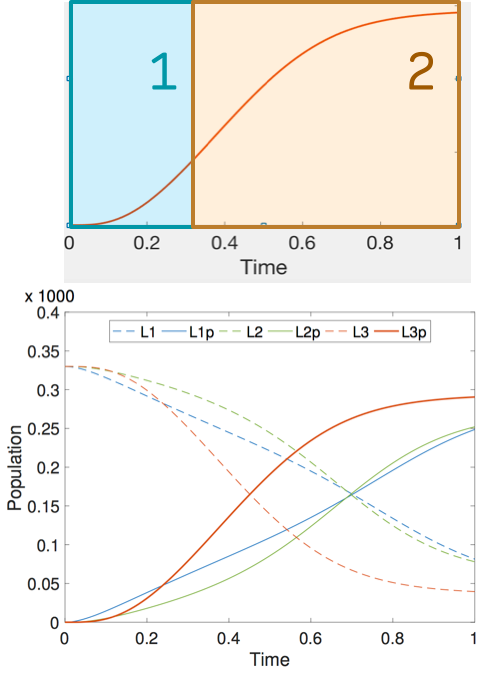
In this project we study bone remodelling, i.e., the biological process of bone renewal, through the use of computational methods and formal analysis techniques. Bone remodelling is a paradigm for many homeostatic processes in the human body and consists of two highly coordinated phases: resorption of old bone by cells called osteoclasts, and formation of new bone by osteoblasts. Specifically, we aim to understand how diseases caused by the weakening of the bone tissue arise as the resulting process of multiscale effects linking the molecular signalling level (RANK/RANKL/OPG pathway) and the cellular level.
To address crucial spatial aspects such as cell localisation and molecular gradients, we developed a modelling framework based on spatial process algebras and a stochastic agent-based semantics, realised in the Repast Simphony tool. This resulted in the first agent-based model and tool for bone remodelling, see also the Tools page.
In addition, we explored probabilistic model checking methods to precisely assess the probability of a given bone disease to occur, and also hybrid approximations to tame the non-linear dynamics of the system.